Background / justification
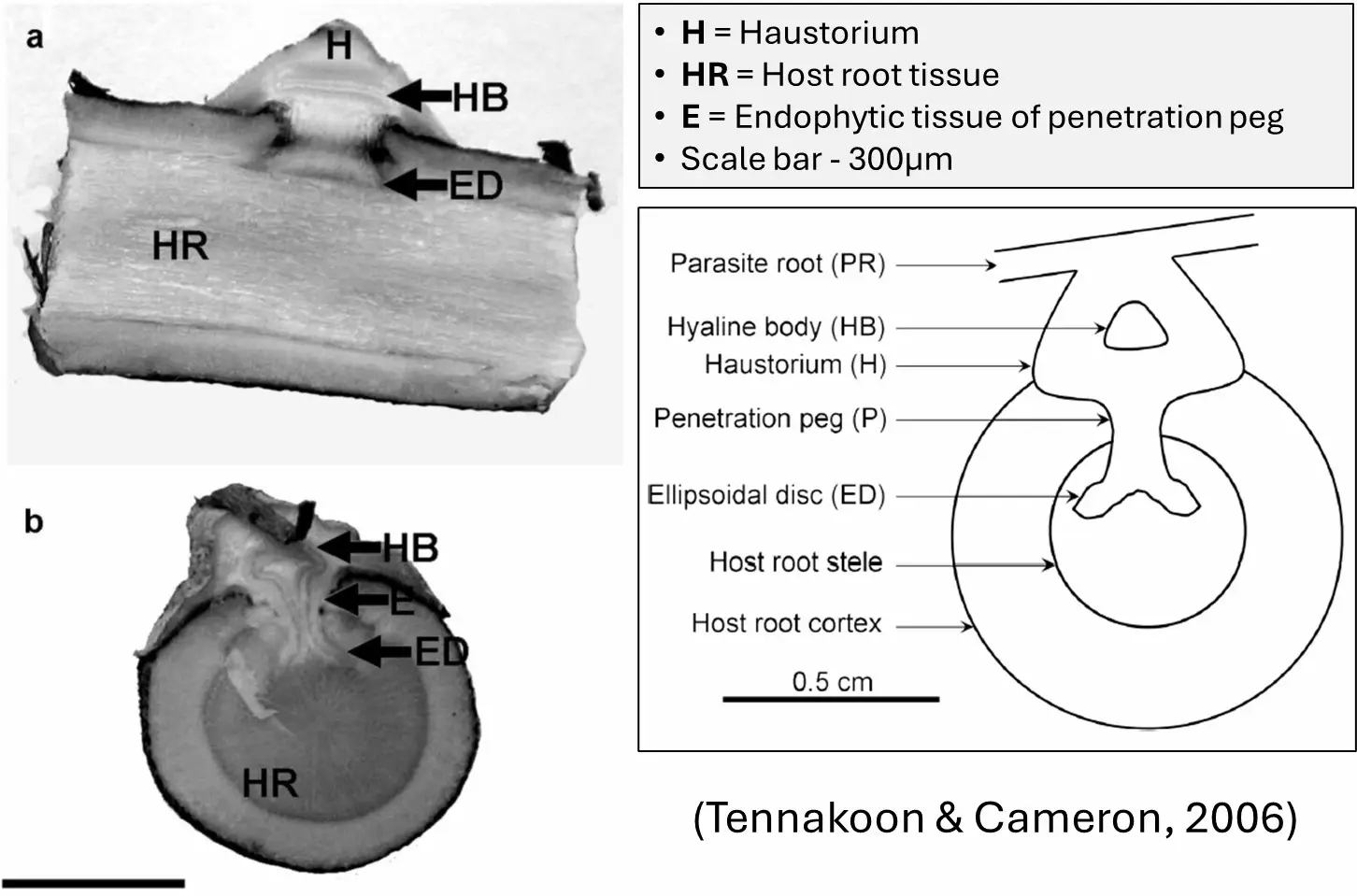
There is significant interest in cultivating ʻiliahi for ecological restoration as well as commercial production. The selection of suitable host species may facilitate the growth of hemiparasitic ʻplants, yet literature addressing host selection for ʻiliahi is limited. N-fixing legumes have been thought to be superior hosts, although that is not true in all instances (Annapurna et al., 2008; Ouyang et al., 2016). The physiological mechanisms of root hemiparasitism in Santalum and the characteristics that define host quality are not well understood. One characteristic of functional haustoria on a high-quality host is the development of elongated “finger” parenchyma cells in the haustoria at the parasite-host cellular interface (Ouyang et al., 2016). Finger parenchyma cells can be present in associations with leguminous and non-leguminous host species and may play a role in effective resource extraction (Tennakoon & Cameron, 2006) (Figure 1). The establishment and maintenance of healthy haustoria connections characterized by finger parenchyma cells may be reliant on specific host root exudate signals (Li et al., 2022) and endogenous biochemical defenses of the ʻiliahi and the host (Mohapatra & Anil 2022).
It is also not completely understood how roots of Santalum species find the roots of their hosts, however, root exudates of high-quality hosts may also contain haustoria-inducing factors such as DMBQ that can act as a chemical signal to guide the growth of parasite roots towards host roots and stimulate the formation of haustoria (Li et al., 2023). The Hawaiʻi endemic S. paniculatum forms well developed haustoria with several Hawaiian native species including, A. koa and M. polymorpha, and its haustoria can also attach to other surfaces besides plants roots including N-fixing nodules of A. koa, fertilizer beads, cinder, and perlite (Figure 2). The haustoria is the site of resource transfer from the host to the parasite, although the transfer of resources in the opposite direction has also been detected in S. album with its hosts in the field (Lu et al., 2020).The midday leaf water potential of Santalum species is observed to be consistently lower than their connected hosts (Světlíková et al., 2018), and this parasite-host water potential gradient is thought to control the direction and potentially the rate of resource transfer between parasite and host. Additionally, the lack of lumen-to-lumen connectivity between the ʻiliahi and host xylem vessels implies that transferred resources must traverse cell membranes, and the activity of membrane-bound transporter proteins may also affect the rate of transfer (Loveys et al., 2001; Těšitel et al., 2015).
The objective of this project is to examine the previously mentioned physiological aspects of root hemiparasitism in S. paniculatum to develop a greater understanding of the parasite-host relationship. We will conduct three experiments that will 1) examine the influence of host pairing on differential expression of genes associated with plant defense response of ʻiliahi and its host, 2) use X-ray CT to assess root architecture and haustoria development in response to host pairing and a potent haustoria inducing factor (DMBQ, 6-dimethoxy-p-benzoquinone), and 3) manipulate the ʻiliahi-host water potential gradient to determine its effect on the rate and direction of resource transfer between the ʻiliahi and its host. This work will ultimately help to inform the host selection process for ʻiliahi and other Santalum species beyond Hawaiʻi.

Project objectives
Experiment 1: Influence of parasitic association on gene expression and biochemical defense responses of root hemiparasite (Santalum paniculatum) and host (Cajanus cajans).
- Obj. 1: Explore changes in gene expression of ʻiliahi and host in response to pairing.
- Obj. 2: Determine whether genes associated with plant biochemical defense response and nutrient transporter proteins are differentially expressed in host pairing.
- Obj. 3: Assess changes in plant biochemical defenses in response to host pairing.
Experiment 2: Root architecture and haustoria development of the root hemiparasite (Santalum paniculatum) in response to host pairing and haustoria-inducing factor (HIF) DMBQ.
- Obj. 1: Use X-ray CT scans to nondestructively monitor root architecture development in response to different native host species and application of the haustoria-inducing factor (HIF) 6-dimethoxy-p-benzoquinone (DMBQ).
- Obj. 2: Determine the limitations of using X-ray CT to detect haustoria connections.
- Obj.3: Determine whether the application of DMBQ can enhance the growth and haustoria connections in native host species pairings.
Experiment 3: Influence of ‘iliahi-host plant water potential gradient on the rate and direction of haustoria borne resource transfer.
- Obj. 1: Determine the influence of the water potential gradient between the ʻiliahi and its host on the degree and direction of resource transfer.
- Obj. 2: Determine if concurrent resource transfer occurs through haustoria connections.
Approach
Experiment 1
Influence of parasitic association on gene expression and biochemical defense responses of root hemiparasite (Santalum paniculatum) and host (Cajanus cajans).
Design
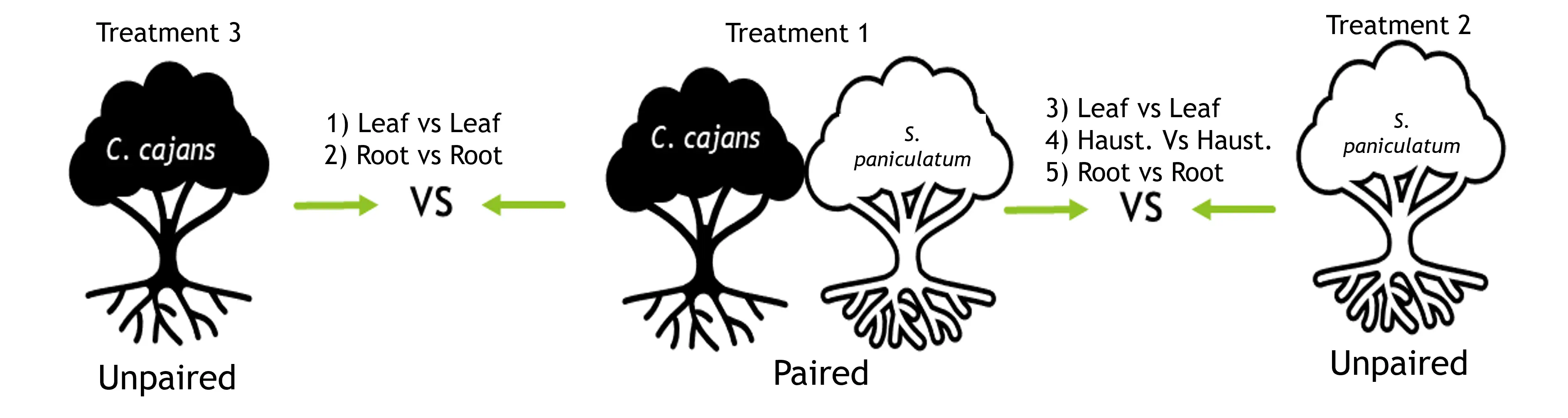
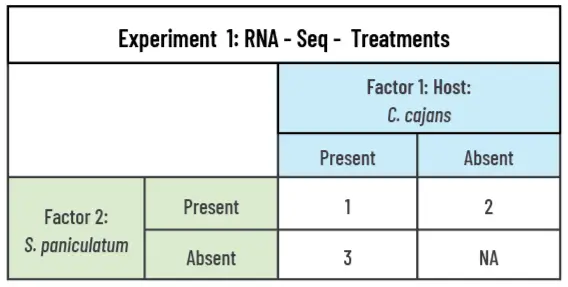
The experiment will be a two-factor design with the S. paniculatum (2 levels: present vs absent) and Host: C. cajans (2 levels: present vs absent) as the predicting factors combined in an incomplete factorial to form 3 treatments (Table 1). For the RNA-seq analysis, RNA will be extracted from three tissue types (1. Haustoria, 2. Fine root, 3. Leaf) for S. paniculatum, and two types (1. Fine root & 2. Leaf) for C. cajans(Figure 3). cDNA libraries generated from the extracted mRNA will be sequenced using HiSeq2000 and sequence reads for S. paniculatum will be aligned to a draft genome of S. album to generate gene counts. The sequence reads for C. cajans will be aligned to a draft genome of C. cajans to generate gene (Zhang et al., 2023)counts. Comparison of gene counts to determine differentially expressed genes will be made between similar tissue types of the paired versus unpaired S. paniculatum and host (Figure 3). Mean gene counts will be calculated across three biological replicates, and each biological replicate will be composed of tissue pooled from 10 individuals. Thirty total cDNA libraries will be sequenced, eighteen (3 biological reps. x 3 tissue types x 2 (paired vs unpaired)) for S. paniculatum and 12 (3 biological reps x 2 tissue types x 2 (paired vs unpaired)) for C. cajans. We will grow 30 parasite-host pairs (container with parasite and host), 30 unpaired S. paniculatum, and 30 unpaired C. cajans. Paired and unpaired host seedlings will be transplanted into designated containers when the S. paniculatum is 6 months old. The parasite-host pair, along with unpaired S. paniculatum and unpaired hosts, will be grown for 12 additional months.
Experiment 1: Assessment
- Obj. 1: We will use the HiSeq2000 platform to generate 100bp paired-end reads of the cDNA libraries generated from the extracted mRNA. Sequence reads for S. paniculatum will be aligned to a S. album reference genome (Zhang et al., 2023) and C. cajans will be aligned with a C. cajans reference genome (Varshney et al., 2012) to generate mean gene counts. Three lists of differentially expressed genes will be generated for the comparison of haustoria, root, and shoot tissue between paired and unpaired S. paniculatum. Two lists of differentially expressed genes will be generated for the comparison of root and shoot tissue between paired and unpaired C. cajans (Figure 3).
- Obj. 2: We will analyze the list of differentially expressed genes to determine whether genes associated with jasmonic acid biosynthesis, salicylic acid biosynthesis, and nutrient transporter protein are influenced by parasitic association.
- Obj. 3: The concentration of jasmonic acid and salicylic acid in three tissue types will be measured using LC-ESI-MS/MS at the metabolite profiling facility at Bindley Science Center. We will measure the activity of superoxide dismutase (SOD) and guaiacol peroxidase (POX), and quantify nonstructural carbohydrates and soluble sugars, total phenols, and total flavonoids in each tissue type. We will incorporate all tissue we collect for the genetic and biochemical defense analyses into the dry mass calculations. Nondestructive measurements of plant morphology (height, ground line diameter, number of leaves, chlorophyll content) will occur monthly throughout the 12-month pairing phase.
Experiment 2
Root architecture and haustoria development of the root hemiparasite (Santalum paniculatum) in response to host pairing and haustoria-inducing factor (HIF) DMBQ.
Design
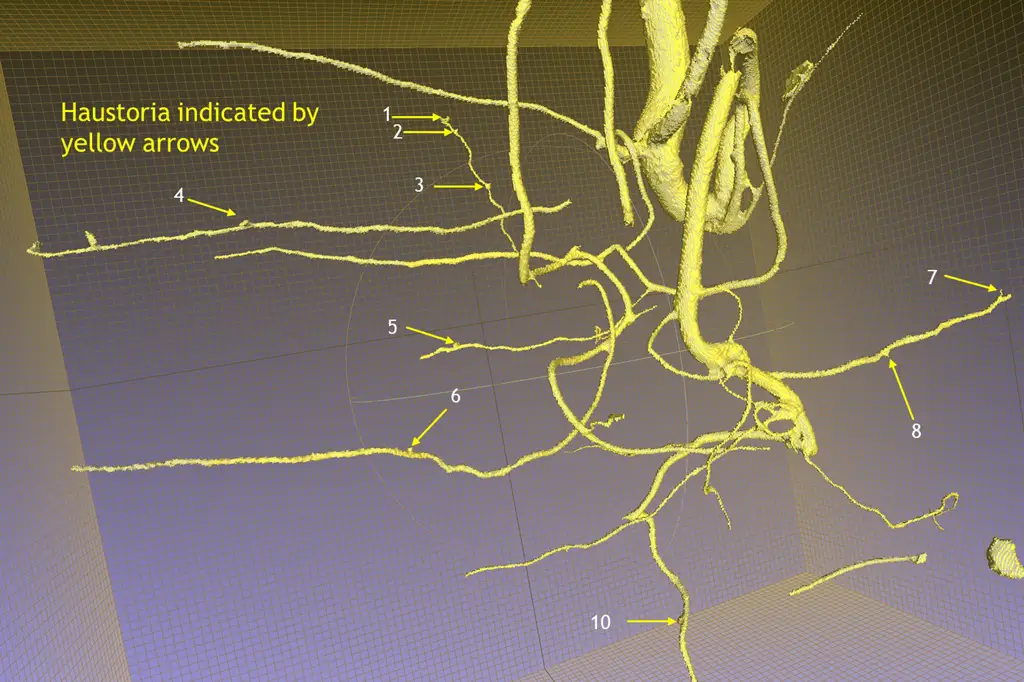
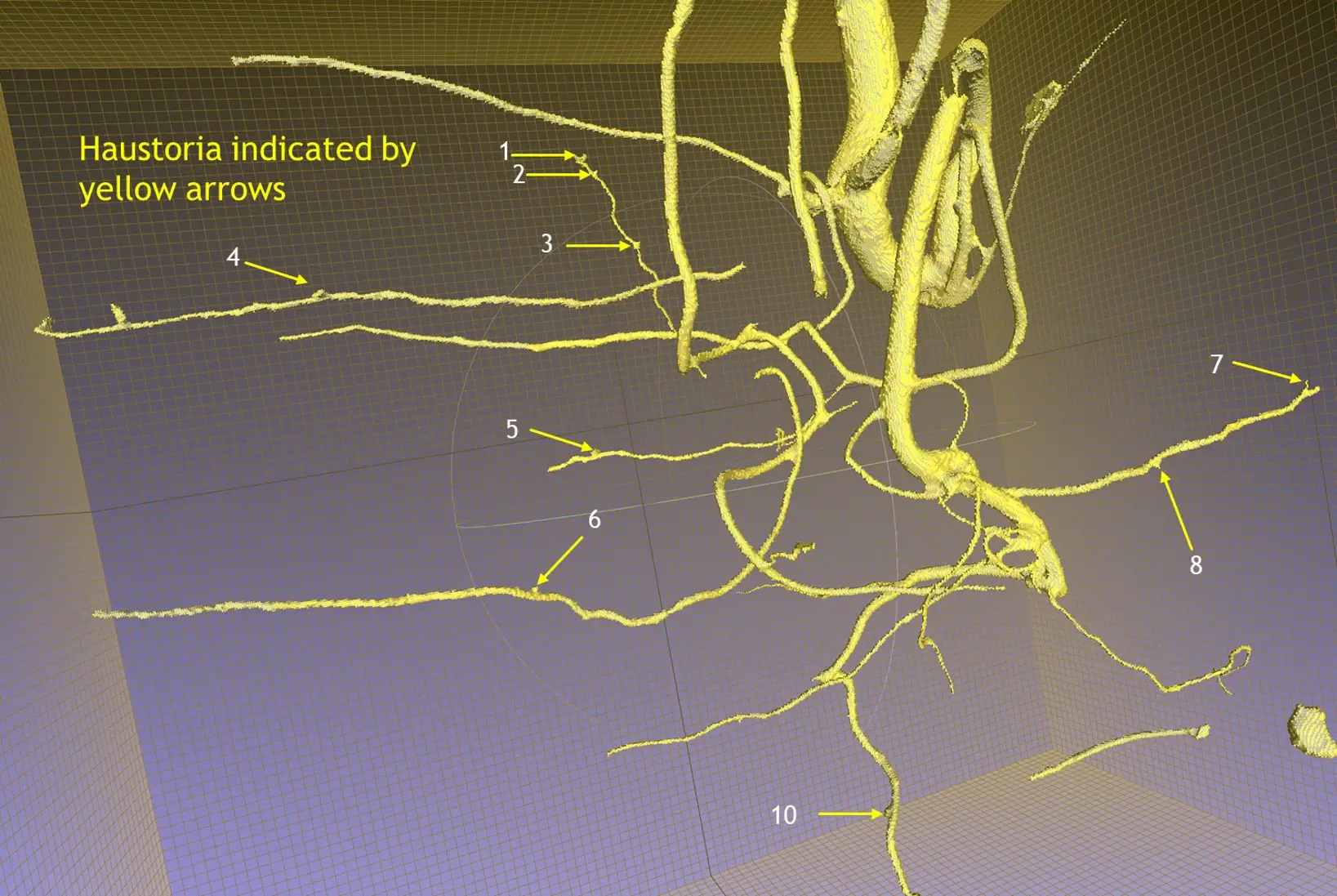
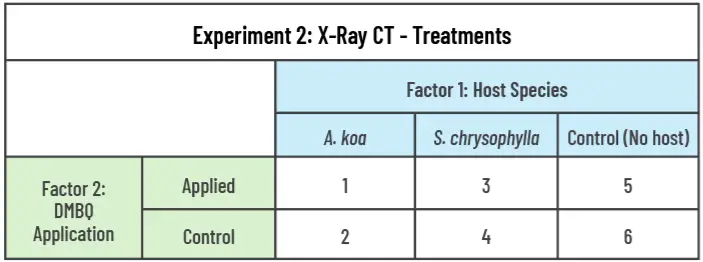
We will use a two-factor design with a full factorial combination of host species (3 levels: A. koa, S. chrysophylla, control) and application of the haustoria-inducing factor, DMBQ (2 levels: Applied vs control) to form 6 unique treatments (Table 2). We will pair twenty-four S. paniculatum seedlings with twenty-four of each host species, and twenty-four S. paniculatum will remain unpaired as a control, resulting in 72 sample containers. Twelve samples from each host species pair and twelve control S. paniculatum (unpaired) will receive a solution of dilute DMBQ, and the remaining half of the samples will not receive any exudates. Samples will be grown together for 12 months in a completely randomized design (CRD) and rearranged monthly. We will use the Purdue Alumni Seed Phenotyping Facility (AAPF) to complete monthly scans of the roots using the X-ray CT scanner (Figure 4) and the shoots to collect RGB imagery and multispectral imagery.
Experiment 2: Assessment
- Obj. 1: On a monthly interval, the root systems of each sample will be nondestructively assessed using X-ray computed tomography (CT). This technology generates a digital twin of the scanned root system, allowing for nondestructive measurement of root volume, root mass, root surface area, root length, and branching angle.
- Obj. 2: We will use X-ray CT to monitor characteristics of haustoria development, including haustoria growth, size of targeted host root, and depth of attachment. We will destructively sample all plants at the end of the grow-out period to assess root and haustoria morphology to compare the values with those from the X-ray CT phenotyping.
- Obj. 2: We will also evaluate morphological responses (Shoot height, number of leaves, leaf surface area, and ground level diameter) at the same interval using high throughput imaging. We will compare mean values for each response (Root: volume, mass, length, surface area, branching direction; Haustoria: number, size, diameter of host root, depth of attachment; Shoot: height, number of leaves, leaf surface area, and ground line diameter) across all treatments to determine the independent and interacting effects of the host species and DMBQ application.
Experiment 3
Influence of ‘iliahi-host plant water potential gradient on the rate and direction of haustoria borne resource transfer.
Design
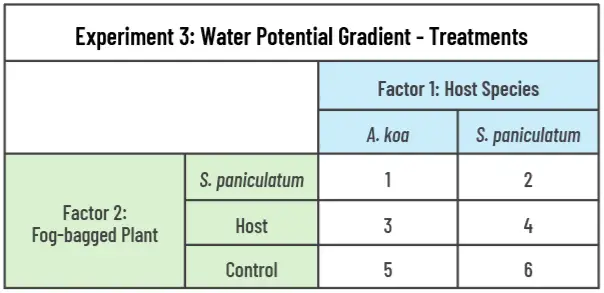
We will use a full factorial design combining host species (2 levels: A. koa and S. paniculatum) and fog-bag treatment (3 levels: host, S. paniculatum, control) to create six unique treatments (Table 3). Sixty S. paniculatum will be grown with A. koa hosts, and sixty will be grown in a pot with another S. paniculatum. Of the 60 S. paniculatum grown with A. koa, 20 will receive the fog-bag treatment on the A. koa, 20 will receive it on the S. paniculatum, and the remaining 20 will not receive the fog-bag treatment. The isotope tracers will be applied to the plant that will receive the fog-bag treatment, and when the fog-bag is not applied, the host will receive the tracers. In the treatment with two S. paniculatum in the same pot, one will be randomly selected to receive the treatment. The pairs will be grown together for 12 months in the same pot, and we will apply a potent haustorium-inducing factor DMBQ to each sample to encourage haustoria formations to form.
After the twelve-month growth out period, we will apply the 13C & 15N stable isotope tracers and collect samples over 8 days as we manipulate the water potential gradient between paired plants. To manipulate the water potential gradient between the S. paniculatum and the host, we will enclose the designated plant in a clear plastic bag suspended by a metal frame and seal the bag at the stem. We will then pump fog into the bag to create a saturated atmospheric condition, which will reduce transpiration and increase water potential while allowing enough light to maintain photosynthesis. Destructive sampling will occur at 0, 2, 4, 6, & 8 days after shading. Four of the twenty samples from each unique treatment will be randomly selected for destructive sampling of the root, stem, and leaf tissue of both plants in the container. All plants will be grown in well-draining sterile sand media, irrigated at high frequency, and drained via vacuum suction to reduce the transfer of 13C & 15N tracers through exudate exchange.
Experiment 3: Assessment
- Obj. 1: Real-time water plant water potential will be recorded during the 8-day experiment period using a passive camera monitoring system that will monitor micro variations in stem swelling as a proxy for water potential. The mean δ15NAir and δ13CVPDB values will be calculated and compared between the S. paniculatum and host in each of the unique treatments. The
- Obj. 2 & 3: Trends in mean δ15NAir and δ13CVPDB for each unique treatment will be analyzed for independent and interacting effects of host species and fog-bag treatment over time. Differences in trends of mean δ15NAir and δ13CVPDB values may indicate that host species or water potential gradients induced by the fog-bag treatment may influence concurrent resource transfer.
Key findings/accomplishments
No data has been collected from these experiments yet.
Future plans
ʻIliahi have already been transplanted for experiment 3, and the host seedlings will be transplanted into containers with S. paniculatum in March 2025. The ‘iliahi seedlings for initially germinated experiments #1 and #2 were used for a pilot trial to practice RNA extraction and refine media for X-ray CT scanning. The new seedlings for experiments 1 and 2 were germinated in March 2025 and will be transplanted into containers in May 2025.
Partners and collaborators
The Hāloa ʻĀina reforestation project provided nursery space for a media pilot trial that was used to refine the media that will be used in the experiment #3. They have also provided all the S. paniculatum seeds for Tawn’s experiments.